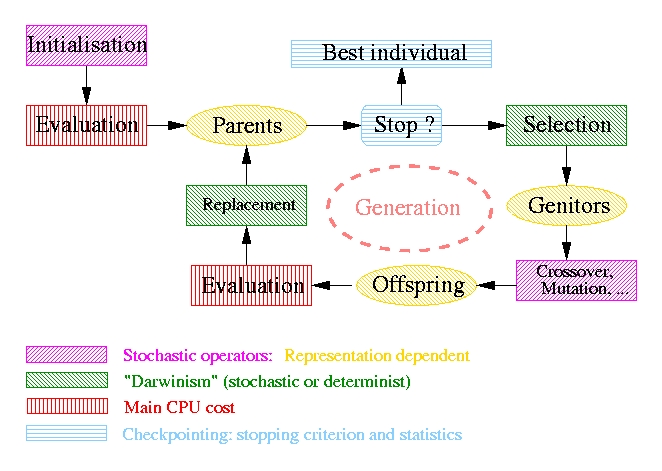
|
//----------------------------------------------------------------------------- // SecondRealEA.cpp //----------------------------------------------------------------------------- //* // Same code than FirstBitEA as far as Evolutionary Computation is concerned // but now you learn to enter the parameters in a more flexible way // (also slightly different than in SecondBitEA.cpp) // and to twidle the output to your preferences (as in SecondBitEA.cpp) // //----------------------------------------------------------------------------- // standard includes #include <stdexcept> // runtime_error #include <iostream> // cout #include <strstream> // ostrstream, istrstream // the general include for eo #include <eo> #include <es.h> |
|
//----------------------------------------------------------------------------- // define your individuals typedef eoReal<eoMinimizingFitness> Indi; |
|
//----------------------------------------------------------------------------- // a simple fitness function that computes the euclidian norm of a real vector // Now in a separate file, and declared as binary_value(const vector<bool> &) #include "real_value.h" |
|
//----------------------------------------------------------------------------- void main_function(int argc, char **argv) { |
|
///////////////////////////// // Fitness function //////////////////////////// // Evaluation: from a plain C++ fn to an EvalFunc Object // you need to give the full description of the function eoEvalFuncPtr<Indi, double, const vector<double>& > plainEval( real_value ); // ... to an object that counts the nb of actual evaluations eoEvalFuncCounter<Indi> eval(plainEval); |
|
//////////////////////////////// // Initilisation of population //////////////////////////////// // Either load or initialize // create an empty pop eoPop<Indi> pop; // create a state for reading eoState inState; // a state for loading - WITHOUT the parser // register the rng and the pop in the state, so they can be loaded, // and the present run will be the exact conitnuation of the saved run // eventually with different parameters inState.registerObject(rng); inState.registerObject(pop); if (loadName != "") { inState.load(loadName); // load the pop and the rng // the fitness is read in the file: // do only evaluate the pop if the fitness has changed } else { rng.reseed(seed); // a Indi random initializer // based on boolean_generator class (see utils/rnd_generator.h) eoUniformGenerator<double> uGen(-1.0, 1.0); eoInitFixedLength<Indi> random(vecSize, uGen); // Init pop from the randomizer: need to use the append function pop.append(popSize, random); // and evaluate pop (STL syntax) apply<Indi>(eval, pop); } // end of initializatio of the population |
|
// sort pop before printing it! pop.sort(); // Print (sorted) intial population (raw printout) cout << "Initial Population" << endl; cout << pop; |
|
///////////////////////////////////// // selection and replacement //////////////////////////////////// |
|
// The robust tournament selection eoDetTournamentSelect<Indi> selectOne(tSize); // is now encapsulated in a eoSelectPerc (entage) eoSelectPerc<Indi> select(selectOne);// by default rate==1 |
|
// And we now have the full slection/replacement - though with // no replacement (== generational replacement) at the moment :-) eoGenerationalReplacement<Indi> replace; |
|
////////////////////////////////////// // The variation operators ////////////////////////////////////// |
|
// uniform chooce on segment made by the parents eoSegmentCrossover<Indi> xoverS; // uniform choice in hypercube built by the parents eoHypercubeCrossover<Indi> xoverA; // Combine them with relative weights eoPropCombinedQuadOp<Indi> xover(xoverS, segmentRate); xover.add(xoverA, hypercubeRate, true); |
|
// offspring(i) uniformly chosen in [parent(i)-epsilon, parent(i)+epsilon] eoUniformMutation<Indi> mutationU(EPSILON); // k (=1) coordinates of parents are uniformly modified eoDetUniformMutation<Indi> mutationD(EPSILON); // all coordinates of parents are normally modified (stDev SIGMA) eoNormalMutation<Indi> mutationN(SIGMA); // Combine them with relative weights eoPropCombinedMonOp<Indi> mutation(mutationU, uniformMutRate); mutation.add(mutationD, detMutRate); mutation.add(mutationN, normalMutRate, true); // The operators are encapsulated into an eoTRansform object eoSGATransform<Indi> transform(xover, pCross, mutation, pMut); |
|
////////////////////////////////////// // termination condition see FirstBitEA.cpp ///////////////////////////////////// eoGenContinue<Indi> genCont(maxGen); eoSteadyFitContinue<Indi> steadyCont(minGen, steadyGen); eoFitContinue<Indi> fitCont(0); eoCombinedContinue<Indi> continuator(genCont); continuator.add(steadyCont); continuator.add(fitCont); |
|
// but now you want to make many different things every generation // (e.g. statistics, plots, ...). // the class eoCheckPoint is dedicated to just that: // Declare a checkpoint (from a continuator: an eoCheckPoint // IS AN eoContinue and will be called in the loop of all algorithms) eoCheckPoint<Indi> checkpoint(continuator); // Create a counter parameter eoValueParam<unsigned> generationCounter(0, "Gen."); // Create an incrementor (sub-class of eoUpdater). Note that the // parameter's value is passed by reference, // so every time the incrementer is updated (every generation), // the data in generationCounter will change. eoIncrementor<unsigned> increment(generationCounter.value()); // Add it to the checkpoint, // so the counter is updated (here, incremented) every generation checkpoint.add(increment); // now some statistics on the population: // Best fitness in population eoBestFitnessStat<Indi> bestStat; // Second moment stats: average and stdev eoSecondMomentStats<Indi> SecondStat; // Add them to the checkpoint to get them called at the appropriate time checkpoint.add(bestStat); checkpoint.add(SecondStat); // The Stdout monitor will print parameters to the screen ... eoStdoutMonitor monitor(false); // when called by the checkpoint (i.e. at every generation) checkpoint.add(monitor); // the monitor will output a series of parameters: add them monitor.add(generationCounter); monitor.add(eval); // because now eval is an eoEvalFuncCounter! monitor.add(bestStat); monitor.add(SecondStat); // A file monitor: will print parameters to ... a File, yes, you got it! eoFileMonitor fileMonitor("stats.xg", " "); // the checkpoint mechanism can handle multiple monitors checkpoint.add(fileMonitor); // the fileMonitor can monitor parameters, too, but you must tell it! fileMonitor.add(generationCounter); fileMonitor.add(bestStat); fileMonitor.add(SecondStat); // Last type of item the eoCheckpoint can handle: state savers: eoState outState; // Register the algorithm into the state (so it has something to save!!) outState.registerObject(parser); outState.registerObject(pop); outState.registerObject(rng); // and feed the state to state savers // save state every 100th generation eoCountedStateSaver stateSaver1(20, outState, "generation"); // save state every 1 seconds eoTimedStateSaver stateSaver2(1, outState, "time"); // Don't forget to add the two savers to the checkpoint checkpoint.add(stateSaver1); checkpoint.add(stateSaver2); // and that's it for the (control and) output |
|
///////////////////////////////////////// // the algorithm //////////////////////////////////////// // Easy EA requires // stopping criterion, eval, selection, transformation, replacement eoEasyEA<Indi> gga(checkpoint, eval, select, transform, replace); // Apply algo to pop - that's it! gga(pop); |
|
// Print (sorted) intial population pop.sort(); cout << "FINAL Population\n" << pop << endl; |
|
} // A main that catches the exceptions int main(int argc, char **argv) { try { main_function(argc, argv); } catch(exception& e) { cout << "Exception: " << e.what() << '\n'; } return 1; } |