


Next: Generalities and file formats
Up: GenRGenS v2.0 User Manual
Previous: Contents
Contents
Random sequences can be
used to extract relevant information from biological sequences. The random
sequences represent the ``background noise'' from which it is possible to differentiate the real
biological information. Random sequences
are widely used to detect over-represented and under-represented motifs,
or to determine whether the scores of pairwise alignments are
relevant. Analytic approaches exist for solving these kinds of problems
(see e.g. [9].) although for the most complex cases, an
experimental approach (i.e. the computer generation of random
sequences) is still necessary.
Some programs are already currently available for generating random
sequences. For example, the GCG package contains a few generation tools,
such as HmmerEmit that generates sequences according to HMM profiles,
and Corrupt that adds random mutations to a given
sequence [3]. Seq-Gen randomly simulates the evolution of
nucleotide sequences along a phylogeny [1]. The Expasy
server has RandSeq, which generates random amino acid
sequences according to a Bernoulli
process [7]. Shufflet is a program that generates random
shuffled sequences [4]. However, until now, there has been no software
package that can integrate several statistical and syntaxical models of
random sequences and combine them. This is the purpose of
GenRGenS.
The random sequence models currently handled by GenRGenS
are the following:
- MARKOV : Markovian random generation. Puts probabilistic constraints on the occurences of
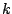 -mers among
the generated sequences. A markovian model can be automatically built from real biological data by a tool
bundled with GenRGenS
-mers among
the generated sequences. A markovian model can be automatically built from real biological data by a tool
bundled with GenRGenS
- GRAMMAR : Random generation based on context-free grammars. This syntaxic formalism allows to take
into account both sequential and structural constraints. Most long-range interactions and correlations
can be captured by this formalism.
- MASTER : Random generation of hierarchical sequences. Combines different levels of abstraction. Sequences of symbols
are generated using a master description file and then some of these symbols are rewrited into sequences generated
with respect to auxiliary description files and distributions over sequences lengths.
- RATEXP : Prosite patterns and rational expressions. Generates sequences uniformly at random from a rational expression
or a prosite pattern. Long ago, searches in language theory stated that these formalisms' expressivity are
included in that of the context-free grammars'1.1. However,
more efficient generation algorithms are available for this subclass. Moreover, support for Prosite patterns allows
a copy/paste approach for random generation that some may find convenient.



Next: Generalities and file formats
Up: GenRGenS v2.0 User Manual
Previous: Contents
Contents
Yann Ponty
2007-04-19